How wiggly spaghetti guard the genome
RESEARCH HIGHLIGHTS
26 April – Tiny pores in the cell nucleus play an essential role in healthy ageing by protecting the DNA from viruses and DNA-damaging substances. A team of scientists in Germany from the Institute of Molecular Biology Mainz, Johannes Gutenberg University Mainz and the Max Planck Institute of Biophysics in Frankfurt am Main has now filled a hole in our understanding of the structure and function of these nuclear pores. The scientists showed how intrinsically disordered proteins in the centre of the pore can form a spaghetti-like mobile barrier that permits important cellular factors to pass, but blocks viruses and other organisms and molecules that can cause disease.
Our DNA is protected within the cell nucleus, which is shielded by a barrier called the nuclear membrane. As the cell’s control centre, the nucleus must be able to exchange important messenger molecules, metabolites or proteins with the rest of the cell. To facilitate this, the nuclear membrane has about 2,000 built-in pores, each consisting of about 1,000 proteins.
For decades, researchers have been fascinated by the 3D structure and function of these nuclear pores, which act as guardians of the genome; substances that are required for controlling the cell are allowed to pass in and out, while viruses and other DNA-damaging substances are blocked from entry. The nuclear pores can therefore be thought of as molecular bouncers, each checking many thousands of visitors per second. Only those who have an entrance ticket are allowed to pass.
How do the nuclear pores manage this enormous task? About 300 of the proteins that make up the pore protrude deep into the central opening, like tentacles. Until now, researchers did not know how these tentacles are arranged and how they repel intruders. This is because these proteins are intrinsically disordered – that is, they lack a defined 3D structure – instead, they are flexible and continuously moving, like spaghetti in boiling water.
Combination of microscopy and computer simulations
As these intrinsically disordered proteins (IDPs) are constantly changing their structure, it is very difficult for scientists to decipher their 3D structure and function. Most experimental techniques that researchers use to visualise proteins only work with a defined 3D structure. Because of this, until now the central region of the nuclear pore has just been represented as a simple hole because it was not possible to determine the structure of the IDPs within the opening.
Now, a team of scientists led by Edward Lemke (Professor of Synthetic Biophysics at Johannes Gutenberg University Mainz and Adjunct Director at the Institute of Molecular Biology Mainz) and Gerhard Hummer (Director of the Max Planck Institute of Biophysics) has used a novel combination of synthetic biology, multidimensional fluorescence microscopy and computer-based simulations to study nuclear pore IDPs in living cells.
“We used modern precision tools to mark several points of the spaghetti-like proteins with fluorescent dyes, which we then excite using light and visualise under the microscope,” Lemke explains. “Based on the glow patterns and duration, we were able to deduce how the proteins must be arranged.” Hummer adds, “We then used molecular dynamics simulations to calculate how the IDPs are spatially organised in the pore, how they interact with each other and how they move. For the first time, we could visualise the gateway into the control centre of human cells.”
A dynamic protein network as a transport barrier
Using the techniques above, the scientists discovered that the protein ‘tentacles’ in the pore behave completely differently from what was previously thought, because they interact with each other and with the molecules attempting to enter, moving perpetually like the aforementioned spaghetti in boiling water. This means that in the centre of the pore, there is no hole, but instead a shield of wiggly, spaghetti-like molecules. Viruses or bacteria are too big to get in through this sieve. However, other large molecules needed in the nucleus can pass, as they carry very specific signals. Such molecules have an entry ticket, whereas others, such as viruses and DNA-damaging molecules, usually do not. “By disentangling the nuclear pore filling, we enter a new phase in nuclear transport research,” adds Martin Beck, collaborator and colleague at the Max Planck Institute of Biophysics. Their findings were published today in the journal Nature.
"Understanding how the pores transport or block cargo will help us to identify how errors occur. After all, some viruses manage to enter the cell nucleus despite the barrier," Hummer sums up. “With our combination of methods, we can now study IDPs in more detail to find why they are indispensable for certain cellular functions, despite being error-prone. In fact, IDPs are found in almost all species, even though they carry the risk of forming aggregates during the ageing process, which can lead to neurodegenerative diseases such as Alzheimer's,” Lemke says. By learning how IDPs function, the researchers aim to develop new drugs or vaccines that can prevent viral infections and help promote healthy ageing.
Further details
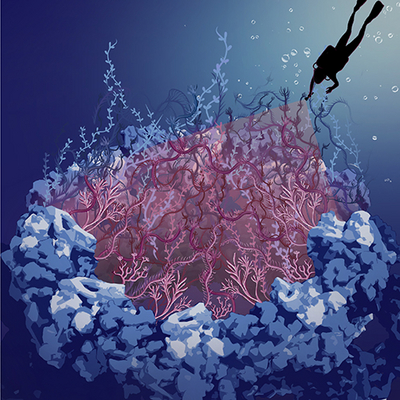
The image shows an artistic impression of the rocky scaffold structure of the nuclear pore complex, filled with intrinsically disordered nucleoporins in the central channel, which are depicted as seaweed. In this work, we “dived” into the dark hole of the nuclear pore complex to shine light on the disordered nucleoporins. Image credit: Sara Mingu.
Further information can be found at www.nature.com/articles/s41586-023-05990-0.
Edward Lemke is an Adjunct Director at IMB and a Professor of Synthetic Biophysics at Johannes Gutenberg University Mainz. Further information about research in the Lemke lab can be found at www.imb.de/lemke.
About the Institute of Molecular Biology gGmbH
The Institute of Molecular Biology gGmbH (IMB) is a centre of excellence in the life sciences that was established in 2011 on the campus of Johannes Gutenberg University Mainz (JGU). Research at IMB focuses on the cutting-edge fields of epigenetics, genome stability, ageing and RNA biology. The institute is a prime example of successful collaboration between a private foundation and government: The Boehringer Ingelheim Foundation has committed 154 million euros to be disbursed from 2009 until 2027 to cover the operating costs of research at IMB. The State of Rhineland-Palatinate has provided approximately 50 million euros for the construction of a state-of-the-art building and is giving a further 52 million in core funding from 2020 until 2027. For more information about IMB, please visit: www.imb.de.
About Johannes Gutenberg University Mainz
Johannes Gutenberg University Mainz (JGU) is a globally recognized research-driven university with around 31,000 students from over 120 nations. Its core research areas are in particle and hadron physics, the materials sciences, and translational medicine. JGU's success in Germany's Excellence Strategy program has confirmed its academic excellence: In 2018, the research network PRISMA+ (Precision Physics, Fundamental Interactions and Structure of Matter) was recognized as a Cluster of Excellence – building on its forerunner, PRISMA. Moreover, excellent placings in national and international rankings as well as numerous honors and awards demonstrate the research and teaching quality of Mainz-based researchers and academics. Further information at www.uni-mainz.de/eng
Boehringer Ingelheim Foundation
The Boehringer Ingelheim Foundation is an independent, non-profit organization that is committed to the promotion of the medical, biological, chemical, and pharmaceutical sciences. It was established in 1977 by Hubertus Liebrecht (1931–1991), a member of the shareholder family of the Boehringer Ingelheim company. Through its Perspectives Programme Plus 3 and its Exploration Grants, the Foundation supports independent junior group leaders. It also endows the international Heinrich Wieland Prize, as well as awards for up-and-coming scientists in Germany. In addition, the Foundation funds institutional projects in Germany, such as the Institute of Molecular Biology (IMB), the department of life sciences at the University of Mainz, and the European Molecular Biology Laboratory (EMBL) in Heidelberg.
Press contact for further information
Dr Ralf Dahm, Director of Scientific Management
Institute of Molecular Biology gGmbH (IMB), Ackermannweg 4, 55128 Mainz, Germany
Phone: +49 (0) 6131 39 21455, Email: press(at)imb.de